Molecular microbiology
Molecular Microbiology
Sepsis and other severe infections are mainly caused by bacteria or fungi. Early identification of the causative pathogen is crucial for effective and successful treatment with antibiotics, antimycotics, supporting medication, and other treatments. Cube Dx molecular pathogen tests support microbiologists and infectious disease specialists in providing early indications of sepsis-causing pathogens in whole blood and other sample matrices.
This is particularly relevant for microorganisms that have been weakened by antimicrobial treatment or which demonstrate slow growth in or on microbiological nutrient media. The broad spectrum of the Cube Dx molecular microbiology test is ideally suited to test for pathogens causing sepsis and other severe infections.
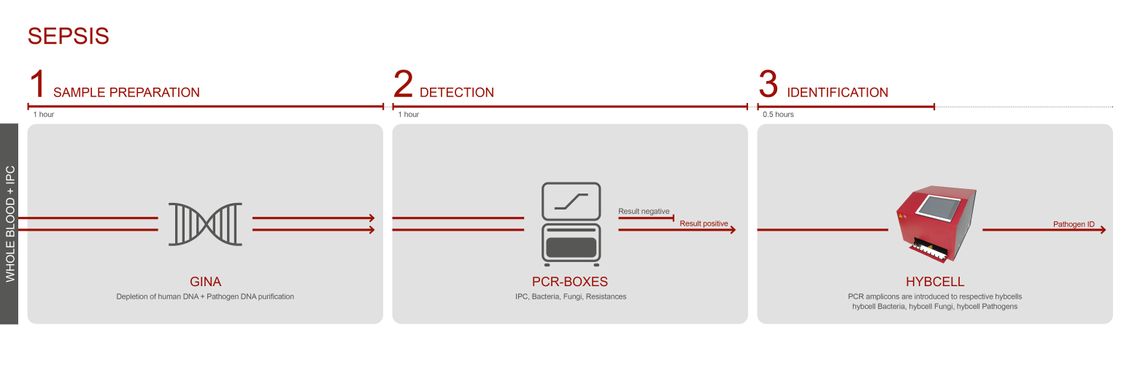
The test is based on three steps: sample preparation and DNA purification, PCR and identification with the hybcell.
The PCR screening for bacteria (16S rDNA) and fungi (28S rDNA) and the follow up identification process on the hybcell are termed compact sequencing, and the test components (PCR mixes and hybcells) and processes are the same for all intended uses. The treatment of the different sample matrices differs strongly between analysing whole blood for sepsis diagnostics or BAL / positive blood cultures for diagnosing pneumonia and other infections.
Whereas analysing whole blood requires the removal of human DNA using the GINA kit, BAL or positive blood cultures require no DNA extraction process whatsoever and use the modulation buffer LINA instead.
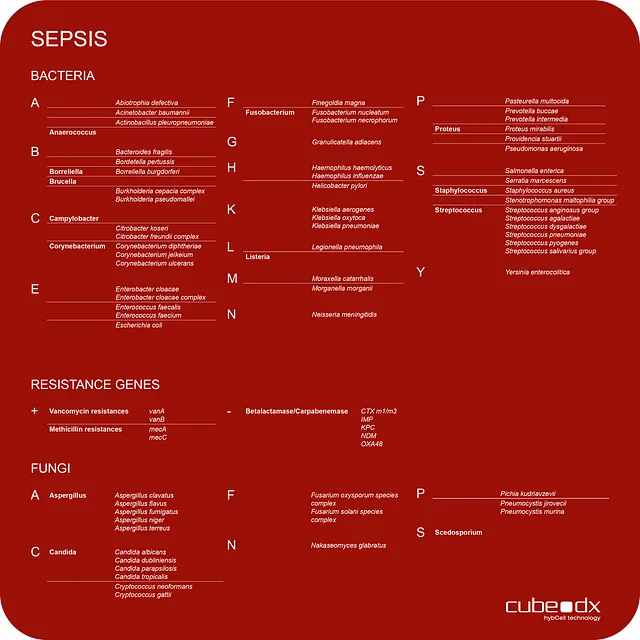
87 microorganisms / resistance genes - in 3 hours
DIAGNOSING SEPSIS – WITHOUT CULTURING / FROM BLOOD
CubeDx supports clinicians by providing a very early indication of sepsis-causing microorganisms directly in whole blood. Testing whole blood reduces the time to result to just 3 hours! Our multiplex technology allows for identification of 87 targets (microorganisms and resistance genes) in parallel.
Fresh EDTA blood (0.5ml) serves as a sample. To validate a negative result, an Internal Process Control (IPC) can optionally be introduced into the sample.
During the GINA process, most cells of human origin are depleted before DNA is purified.
The enriched microbial DNA is amplified using a choice of PCR master-mixes: pan-bacterial PCR, pan-fungal PCR or PCR to screen for several resistance genes.
Positive samples are transferred into the hybcell cartridges, and the microorganisms and resistance genes are identified by the hybcell and its compact sequencing process, with the results subsequently presented in a comprehensive report.
Features / Advantages:
-
Results in just 3 hours!
-
Yield of up to 98% of microbial DNA
-
Limit of detection (whole blood): 20 CFU/mL
-
Clinical sensitivity: 74% and specificity: 98%
-
Early and comprehensive results from 0.5 mL whole blood
-
Sensitive and parallel identification of bacteria, fungi, and resistance genes
-
Identification of multiple pathogens/mixed infections
-
Simple handling, an optimized workflow with 24/7 availability and immediate results
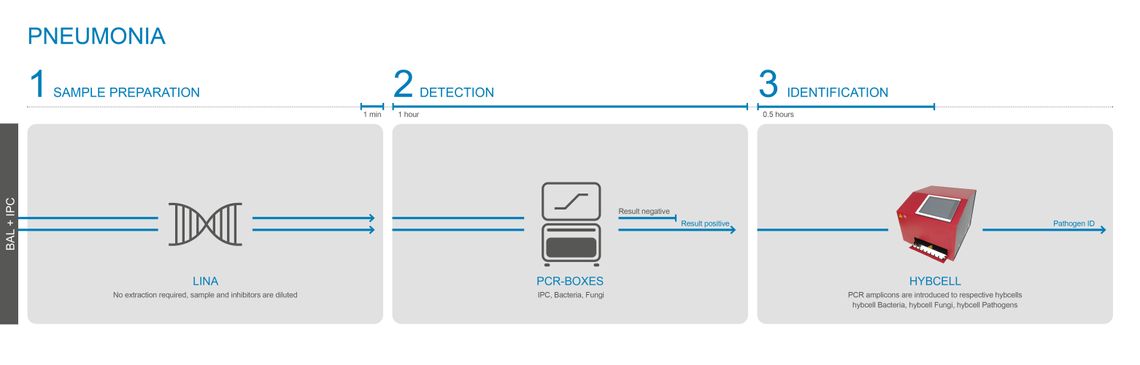
31 microorganisms - in 2 hours
DIAGNOSING PNEUMONIA – WITHOUT EXTRACTION / FROM BAL
Cube Dx’s highly sensitive PCR mixes can test BAL without the need for prior DNA extraction. A simple modulation buffer – LINA – is used to modulate and homogenize BAL. Using the extraction-free protocol shortens the time to result to only 2 hours.
The LINA modulation buffer is used for samples with a relatively high concentration of pathogens, for example BAL. Only a few µL of BAL are added to the ready-to-use LINA tube and the solution is mixed. The mixture is directly used for the follow-up PCR. The test (PCR and hybcell) is the same product as used for testing for sepsis – a software option limits the output to the CE-IVD certified panel for pneumonia.
The LINA transfer and modulation buffer shorten the time for molecular identification to 2 hours – as it eliminates the DNA extraction process.
Features / Advantages:
-
No DNA extraction
-
Identification of pathogens in less than 2 hours
-
Simple and fast
-
Sensitive and parallel identification of bacteria and fungi
-
Identification of multiple pathogens/mixed infections
100 microorganisms / resistance genes - in 2 hours
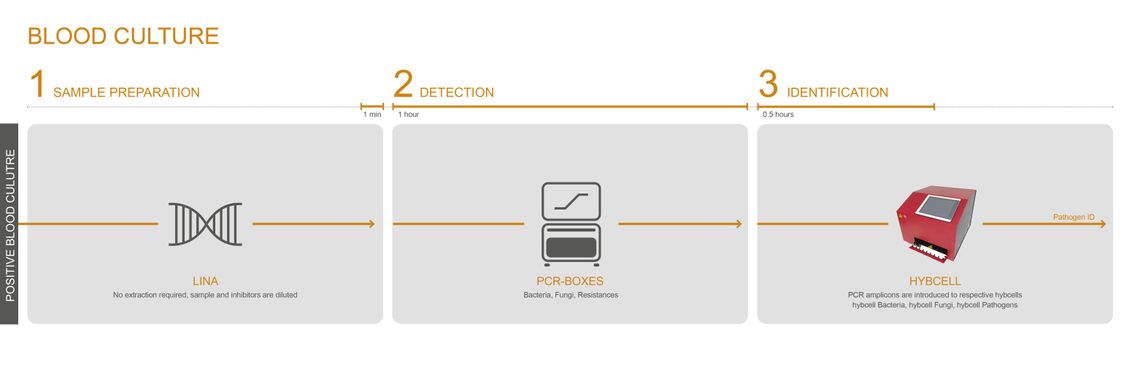
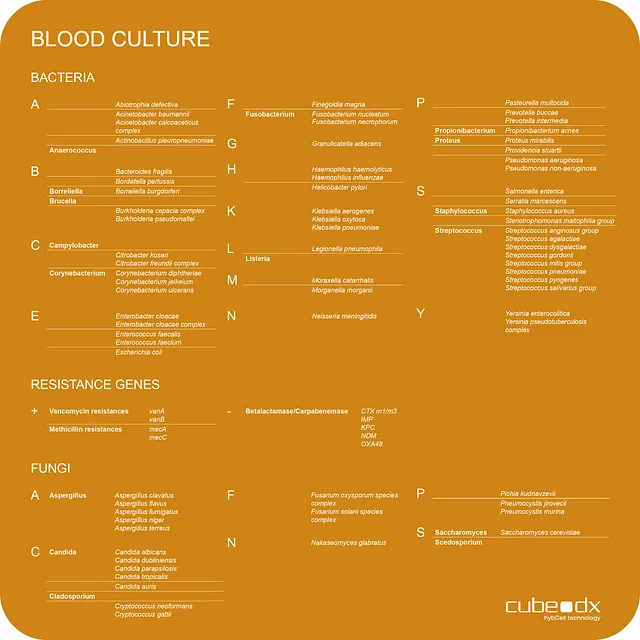
MICROBIAL IDENTIFICATION – DIRECTLY FROM POSITIVE HEMOCULTURE
CubeDx offers fast DNA-based identification of microorganisms grown in blood cultures. No further pure culture is needed, and even slow growers can be identified in parallel to the dominating microorganism. This test can, at the very least, serve as a backup system to identification with MALDI-TOF.
The LINA buffer offers a simple, fast and efficient path to identifying disease-causing pathogens with only 1 minute actual hands-on time. A few µL of positive blood culture can be directly added to the LINA buffer without the need for pure cultures. The mixture is transferred into the PCR module, and identification with hybcell follows with the result being delivered in 2 hours.
Features / Advantages:
-
Bacteria: 56 species + 11 genera
-
Fungi: 19 species + 5 genera
-
9 antibiotic resistance genes
-
Results for mixed infections
-
No DNA extraction
-
Identification of pathogens in less than 2 hours
-
Simple and fast
-
Identification of multiple pathogens/mixed infections
16 fungal genera / 50 fungal species - in 2.5 hours
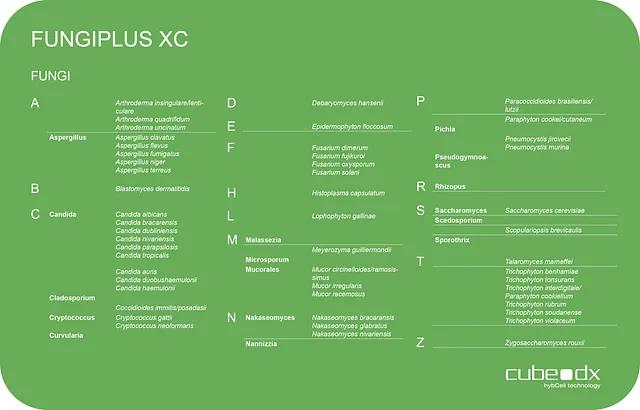
FOR RESEARCH PURPOSES - WIDE MOLECULAR FUNGUS TEST
Cube Dx offers fast and comprehensive DNA-based identification of fungi from different samples for research purposes. By using our GINA sample preparation, the overall time to result for ‘difficult’ samples such as skin or nail scales can also be drastically reduced.
For example, 0.5 mL of fresh EDTA blood can be used as a sample, which is prepared using the GINA sample preparation system. However, any other sample - from BAL to urine, CSF, tissue, skin, nails or hair - can also be used. The sample DNA contained in the eluate serves as the input material for the test.
The microbial DNA is amplified with the PCR master mix and positive samples are transferred to the hybcell cartridges and the fungi are identified by compact sequencing. The results are automatically summarised in a compact report.
Features / Advantages:
-
16 fungal genera
-
50 fungal species
-
66 fungal targets in 2.5 hours
-
Testing of different sample matrices
-
Automated with little hands-on
-
Identification of multiple pathogens/mixed infections
Molecular microbiology - Literature
Available literature
Available literature:
- 2016: Sensitive Molecular Identification of Pathogens causing Implant and Tissue Infections (ITI)
- 2020: Molecular pathogen identification and resistance gene detection from positive blood cultures
- 2020: Presence and distribution of fungal species and dermatophytes in nail and skin samples
- 2021: SMARTDIAGNOS - next generation molecular sepsis diagnosis technology for whole blood samples
- 2022: Extraction-free Molecular Identification of Bacteria/Fungi Directly from BAL Samples
- 2023: Comparison of two molecular assays and MALDI-TOF MS Sepsityper for the rapid identification of pathogens from positive blood cultures
- 2023: Detection of bacterial pneumonia pathogens in patients with COVID-19
- 2023: Diagnostic value of blood culture independent molecular tests for the diagnosis of bloodstream infections in neutropenic patients with fever